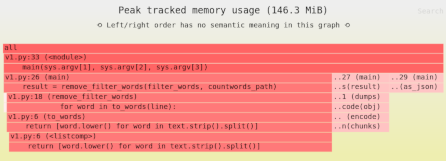
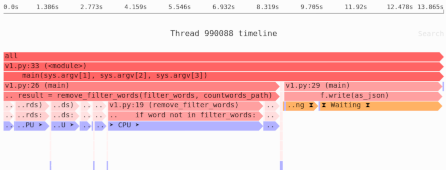
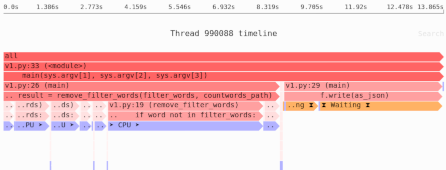
Loading NumPy arrays from disk: mmap() vs. Zarr/HDF5
If your NumPy array is too big to fit in memory all at once, you can process it in chunks: either transparently, or explicitly loading only one chunk at a time from disk. Either way, you need to store the array on disk somehow.
For this particular situation, there are two common approaches you can take:
mmap(), via thenumpy.memmap()API, which lets you treat a file on disk transparently as if it were all in memory.- Zarr and HDF5, a pair of similar storage formats that let you load and store compressed chunks of an array on demand.
Each approach has different strengths and weaknesses, so in this article I’ll explain how each storage system works and when you might want to use each. In particular, I’ll be focusing on data formats optimized for running your calculations, not necessarily for sharing with others.
What happens when you read or write to disk?
When you read a file from disk for the first time the operating system doesn’t just copy the data into your process. First, it copies it into the operating system’s memory, storing a copy in the “buffer cache”.
Why is this useful?
The operating system keeps this buffer cache around in case you read the same data from the same file again.
If you read again, the second read will come from RAM and be orders of magnitude faster.
If you need that memory for something else, the buffer cache will be automatically cleared.
When you write, the data flows in the opposite direction—initially it only gets written to the buffer cache. This means writes are typically quite fast, because you don’t have to wait for the much slower write to the disk, you’re only writing to RAM.
Eventually the data gets flushed from the buffer cache to your hard drive.
mmap()ing an array
For our purposes, mmap() lets you treat a file on disk as if it were an in-memory array.
The operating system will transparently read/write either the buffer cache or the disk, depending on whether the particular part of the array is cached in RAM or not:
- Is the data in the buffer cache? Great, you can just access it directly.
- Is the data only on disk? Access will be slower, but you don’t have to think about it, it’ll get loaded transparently.
As an added bonus, in most cases the buffer cache for the file is embedded into your program’s memory, so you’re not making an extra copy of the memory out of the buffer cache and into your program’s memory.
NumPy has built-in support for mmap():
import numpy as np
array = np.memmap("mydata/myarray.arr", mode="r",
dtype=np.int16, shape=(1024, 1024))
Run that code, and you’ll have an array that will transparently either return memory from the buffer cache or read from disk.
The limits of mmap()
While mmap() can work quite well in some circumstances, it also has limitations:
- The data has to be on the filesystem. You can’t load data from a blob store like AWS S3.
- If you’re loading enough data, reading or writing from disk can become a bottleneck. remember, disks are much slower than RAM. Just because disk reads and writes are transparent doesn’t make them any faster.
- If you have an N-dimensional array you want to slice along different axes, only the slice that lines up with the default structure will be fast. The rest will require large numbers of reads from disk.
To expand on the last item: imagine you have a large 2D array of 32-bit (4 byte) integers, and reads from disk are 4096 bytes. If you read in the dimension that matches the layout on disk, each disk read will read 1024 integers. But if you read in the dimension that doesn’t match the layout on disk, each disk read will give you only 1 relevant integer. You may need 1000× more reads to get the same amount of relevant data!
Zarr and HDF5
To overcome these limits, you can use Zarr or HDF5, which are fairly similar:
- HDF5, usable in Python via
pytablesorh5py, is an older and more restrictive format, but has the benefit that you can use it from multiple programming languages. - Zarr is much more modern and flexible, but has less extensive supports outside Python; Z5 is an implementation for C++. My feeling is that it’s a better choice in most situations unless you need HDF5’s multi-language support, for example Zarr has much better threading support.
For the rest of the article I’ll just talk about Zarr, but if you’re interested there’s a in-depth talk by Joe Jevnik on the differences between Zarr and HDF5.
Using Zarr
Zarr lets you store chunks of data and load them into memory as arrays, and write back to these chunks as arrays.
Here’s how you might load an array with Zarr:
>>> import zarr, numpy as np
>>> z = zarr.open('example.zarr', mode='a',
... shape=(1024, 1024),
... chunks=(512,512), dtype=np.int16)
>>> type(z)
<class 'zarr.core.Array'>
>>> type(z[100:200])
<class 'numpy.ndarray'>
Notice that until you actually slice the object, you don’t get a numpy.ndarray: the zarr.core.Array is just some metadata, you only load from disk the subset of data you slice.
Why Zarr?
Zarr addresses the limits of mmap() we discussed earlier:
- You can store the chunks on disk, on AWS S3, or really in storage system that provides key/value lookup.
- Chunk size and shape are up to you, so you can structure them to allow efficient reads across multiple axes. This is also true of HDF5.
- Chunks can be compressed; this is also true of HDF5.
Let’s go over the last two points.
Chunk dimensions
Let’s say you’re storing a 30000x30000 array. If you want to read in both X and Y axes you can store chunks in the following layout (in practice you’d probably want more than 9 chunks):
Now you can load in either X or Y dimension efficiently: you could load chunks (1, 0), (1, 1), (1, 2), or you could load chunks (0, 0), (0, 1), (0, 2) depending which axis was relevant to your code.
Compression
Each chunk can be compressed. That means you can load data faster than the disk bandwidth: if your data compression ratio is 3×, you can load data 3× as fast as the disk bandwidth, minus the CPU time spent decompressing the data.
Once the chunks are loaded they can be dropped from your program’s memory.
Note: Whether or not any particular tool or technique will help depends on where the actual memory bottlenecks are in your software.
Need to identify the memory and performance bottlenecks in your own Python data processing code? Try the Sciagraph profiler, with support for profiling both in development and production macOS and Linux, and with built-in Jupyter support.
mmap() or Zarr?
So which one should you use?
mmap() is useful when:
- You have multiple processes reading the same parts of the same file. The processes will be able to share the buffer cache via
mmap(), so you just need one copy in memory no matter how many processes are running. - You don’t want to manually manage memory, but just rely on the OS to transparently and automatically do it for you.
Zarr is useful when:
- You’re loading data from some remote source, rather than the local filesystem.
Zarr and HDF5 are useful when:
- Reading from disk is likely to be a bottleneck; compression will give your more effective bandwidth.
- If you need to slice multi-dimensional data across different axes, which they allow by using an appropriate chunk shape and size.
As a first pass, I would personally default to Zarr, just because of the flexibility it gives you.