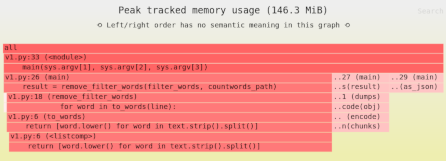
Reducing Pandas memory usage #1: lossless compression
You’re loading a CSV into Pandas, and it’s using too much RAM: your program crashes if you load the whole thing. How do you reduce memory usage without changing any of your processing code?
In this article I’ll show you how to reduce the memory your DataFrame uses at the time it is initially loaded, using four different techniques:
- Dropping columns
- Lower-range numerical dtypes.
- Categoricals.
- Sparse columns.
Technique #1: Don’t load all the columns
Quite often the CSV you’re loading will include columns you don’t actually use when processing the data. If you don’t use them, there’s no point in loading them!
In the following example, I am loading the full CSV, checking how much memory is used by the DataFrame, and then shrinking down to just the columns I’m interested in:
>>> import pandas as pd
>>> df = pd.read_csv("voters.csv")
>>> df.info(verbose=False, memory_usage="deep")
...
memory usage: 71.2 MB
>>> df = df[["First Name ", "Last Name "]]
>>> df.info(verbose=False, memory_usage="deep")
...
memory usage: 8.3 MB
Peak memory usage is 71MB, even though we’re only really using 8MB of data. Note that these usage numbers are somewhat inaccurate; the important thing is the ratio.
But since we know in advance we only need those two columns, we don’t need to load everything, we can just load only the columns we care about, thus reducing peak memory usage to 8MB:
>>> df = pd.read_csv(
... "voters.csv", usecols=["First Name ", "Last Name "])
>>> df.info(verbose=False, memory_usage="deep")
...
memory usage: 8.3 MB
Technique #2: Shrink numerical columns with smaller dtypes
Another technique can help reduce the memory used by columns that contain only numbers.
Each column in a Pandas DataFrame is a particular data type (dtype).
For example, for integers there is the int64 dtype, int32, int16, and more.
Why does the dtype matter? First, because it affects what values you can store in that column:
int8can store integers from -128 to 127.int16can store integers from -32768 to 32767.int64can store integers from -9223372036854775808 to 9223372036854775807.
Second, the larger the range, the more memory is used.
For example, int64 uses 4× as much memory as int16, and 8× as much as int8.
By default when Pandas loads a CSV, it guesses at the dtypes.
If it decides a column volumes are all integers, by default it assigns that column int64 as the dtype.
As a result, if you know that the numbers in a particular column will never be higher than 32767, you can use an int16 and reduce the memory usage of that column by 75%.
And if the values will never be higher than 127, you can use an int8, using even less memory.
For example, here we see how much memory a column uses (the nbytes attribute) when loaded with the default int64 vs. when we load it with int8.
We know that int8 won’t lose any data because the numbers in the column only range from 1 to 11:
>>> df = pd.read_csv("voters.csv")
>>> df["Ward Number "].memory_usage(index=False, deep=True)
550688
>>> df["Ward Number "].max()
11
>>> df["Ward Number "].min()
1
>>> df = pd.read_csv("voters.csv", dtype={"Ward Number ": "int8"})
>>> df["Ward Number "].memory_usage(index=False, deep=True)
68836
Notice how much less memory the column is using.
Technique #3: Shrink categorical data using Categorical dtypes
What about non-numerical data? In some cases you can shrink those columns as well.
In the voter registration database I am using as an example there is a column for party affiliation, by default parsed as a string, with only a fixed number of values in it. Political parties don’t get formed overnight, after all.
>>> set(df["Party Affiliation "])
{'Q ', 'S ', 'L ', 'R ', 'H ', 'BB', 'D ', 'K ', 'O ', 'X ', 'A ', 'Z ', 'EE', 'F ', 'P ', 'G ', 'T ', 'CC', 'J ', 'AA', 'Y ', 'U '}
>>> df["Party Affiliation "].memory_usage(index=False, deep=True)
4061324
Even though the values are limited to a specific set, they are still stored as arbitrary strings, which have overhead in memory.
Imagine a gender column that only says "FEMALE", "MALE", and "NON-BINARY" over and over again—that’s a lot of memory being used to store the same three strings.
A more compact representation for data with only a limited number of values is a custom dtype called Categorical, whose memory usage is tied to the number of different values.
When we load the CSV, we can specify that a particular column uses this dtype, and the resulting memory usage is much smaller, 69KB instead of 550KB:
>>> df = pd.read_csv(
... "voters.csv", dtype={"Party Affiliation ": "category"})
>>> df["Party Affiliation "].head()
0 U
1 U
2 U
3 R
4 U
Name: Party Affiliation , dtype: category
Categories (22, object): [A, AA, CC, D, ..., Y, Z, BB, EE]
>>> df["Party Affiliation "].memory_usage(index=False, deep=True)
70774
That’s much smaller.
Note: Whether or not any particular tool or technique will help depends on where the actual memory bottlenecks are in your software.
Need to identify the memory and performance bottlenecks in your own Python data processing code? Try the Sciagraph profiler, with support for profiling both in development and production macOS and Linux, and with built-in Jupyter support.
Technique #4: Sparse series
If you have a column with lots of empty values, usually represented as NaNs, you can save memory by using a sparse column representation.
It won’t waste memory storing all those empty values.
For example, three quarters of this column are empty values:
>>> df = pd.read_csv("voters.csv")
>>> series = df["Mailing Address - Apartment Number "]
>>> series.memory_usage(index=False, deep=True)
2623975
>>> len(series)
68836
>>> len(series.dropna())
13721
We can convert the series to a sparse series, and then it’ll use less memory:
>>> sparse_series = series.astype("Sparse[str]")
>>> len(sparse_series)
68836
>>> sparse_series.memory_usage(index=False, deep=True)
2237939
In this case the memory savings aren’t huge (2.6MB → 2.2MB), but there is some saving; for integers or other number-like types the savings would be more significant.
Unfortunately it’s not yet possible to use read_csv() to load a column directly into a sparse dtype.
Compression is your friend
As you’ve seen, simply by changing a couple of arguments to pandas.read_csv(), you can significantly shrink the amount of memory your DataFrame uses.
Same data, less RAM: that’s the beauty of compression.
Need even more memory reduction? You can use lossy compression or process your data in chunks.