Why Polars uses less memory than Pandas
Processing large amounts of data with Pandas can be difficult; it’s quite easy to run out of memory and either slow down or crash. The Polars dataframe library is a potential solution.
While Polars is mostly known for running faster than Pandas, if you use it right it can sometimes also significantly reduce memory usage compared to Pandas. In particular, certain techniques that you need to do manually in Pandas can be done automatically in Polars, allowing you to process large datasets without using as much memory—and with less work on your side!
This does require you to use the correct Polars APIs. And it won’t solve all your problems, even if it does make your life easier.
In this article we’ll:
- See how we can optimize Pandas memory usage with a little work.
- See how Polars can, in some cases, use these techniques automatically.
- Note at least some of the ways you will need to intervene manually to reduce memory usage.
An example: going from naive Pandas to memory-optimized Pandas
To help understand the way Polars can help reduce memory usage compared to Pandas, we’ll start with a concrete example and implement it in Pandas. We are going to look at recorded bus route times for the Boston area’s transit authority, the MBTA, and try to find bus routes that are extra slow.
We’ll use data from 2022, available here in the form of CSVs. Each month’s data is a single CSV of about 300MB; we’ll be looking at the data from May.
First we’ll implement a particular query with a naive Pandas implementation, and then with a more optimized but still Pandas implementation. In the next major section we’ll switch to Polars.
Step 1: A more efficient memory representation and file format
Here’s a sample of the data, with some of the columns omitted:
| service_ date | route_ id | direction_ id | standard_ type | scheduled | scheduled_ headway | headway |
|---|---|---|---|---|---|---|
| 2022-05-01 | “01” | “Inbound” | “Schedule” | “1900-01-01 06:05:00.000” | NA | NA |
| 2022-05-01 | “01” | “Inbound” | “Schedule” | “1900-01-01 06:25:00.000” | NA | NA |
| 2022-05-01 | “01” | “Inbound” | “Headway” | “1900-01-01 06:25:00.000” | “1200” | “841” |
| 2022-05-01 | “01” | “Inbound” | “Schedule” | “1900-01-01 06:29:00.000” | NA | NA |
| 2022-05-01 | “01” | “Inbound” | “Schedule” | “1900-01-01 06:30:00.000” | NA | NA |
Inbound vs Outbound means going towards or from Boston, a city which is also known as “The Hub”.
The first thing to notice is that many of these columns can be represented using a more memory-efficient data type, without losing any information. See this article on reducing Pandas memory usage with compression for more details.
- Instead of strings,
service_date,actualandscheduled_headwaycan be timestamps. In the real world I’d do more work to ensure they’re actually dates or time-of-days, but for the limited example we’ll be working on a timestamp will suffice. - Instead of strings,
route_id,direction_id, andstandard_typecan be categoricals.
We may wish to process the file multiple times, for example to try different queries. In that case we won’t want data type conversion to happen after loading, we’d ideally have the data stored on disk in a way that remembers the data types we’d like to use. CSV does not qualify, it’s basically just a pile of strings.
In addition, loading a CSV can be slow, with lots of parsing involved. A better alternative to CSVs is the Parquet data format: it has an actual concept of data types that is similar to Pandas, and it is faster to load.
As our first step, then, we will load the CSV, choose better column types, and write the result to a Parquet file:
import sys
import pandas as pd
df = pd.read_csv(
sys.argv[1],
dtype={
"route_id": "category",
"direction_id": "category",
"point_type": "category",
"standard_type": "category",
},
parse_dates=["service_date", "scheduled", "actual"],
)
df.to_parquet(sys.argv[1].replace(".csv", ".parquet"))
As an added bonus, Parquet uses compression: the new file is 20MB, compared to 300MB for the CSV. Keep in mind this is just the on-disk size. The data will have to be uncompressed before loading into memory, so on-disk compression doesn’t help with memory usage.
Step 2: Finding slow bus routes with a naive Pandas implementation
In order to find slow bus routes, we’re going to focus on “headways”: how often a particular bus route arrives.
If we look at the sample data above, we can see that inbound bus 1 is supposed to arrive every 1200 seconds, but on May 1st it actually arrived more quickly, with a difference of 841 seconds.
Not all rows have headway information; we only want rows where standard_type is Headway.
Here’s our algorithm:
- Get rid of all rows that aren’t headway information.
- Calculate the ratio of actual headway to expected headway; if it’s bigger than 1, that means the bus arrived late.
- For every pair of route number and direction (inbound/outbound), pick the median headway ratio for the month.
- Find the 5 route pairs with the worst median ratio.
This is probably a bad way to find slow buses, but we’re just using this as an example, so that’s fine.
Here’s a first pass, a naive implementation:
import pandas as pd
def find_worst_headways():
# Load the data:
data = pd.read_parquet("MBTA-2022-05.parquet")
# Filter down to headway points only:
data = data[data["standard_type"] == "Headway"]
# Calculate ratio of actual headway to expected headway:
data["headway_ratio"] = (
data["headway"] / data["scheduled_headway"]
)
# Group by route and direction (Inbound/Outbound):
by_route = data.groupby(["route_id", "direction_id"])
# Find median headway ratio for each route:
median_headway = by_route[["headway_ratio"]].median()
# Return the worst 5 routes:
return median_headway.nlargest(
5, columns=["headway_ratio"]
)
print(find_worst_headways())
Here’s the result:
headway_ratio
route_id direction_id
108 Outbound 2.900000
88 Outbound 1.680000
83 Outbound 1.565000
134 Outbound 1.431111
Inbound 1.346667
For some reason one route ID is blank. I haven’t investigated why because this is just an example, but perhaps the input data is malformed. As long as our later implementations give the same results, this doesn’t really matter for this article, we’re comparing apples to apples. In the Polars implementation it comes out as 134, with all other results being the same.
By running the program using /usr/bin/time -v, we can see max RSS (resident) memory usage, and wallclock time and CPU time:
User time (seconds): 0.84
System time (seconds): 1.33
Percent of CPU this job got: 491%
Elapsed (wall clock) time (h:mm:ss or m:ss): 0:00.44
Maximum resident set size (kbytes): 909500
Pandas isn’t parallelized, but the Parquet loading library (in this case Arrow) can take advantage of multiple CPUs.
Step 3: Investigating memory usage
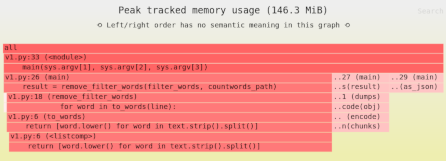
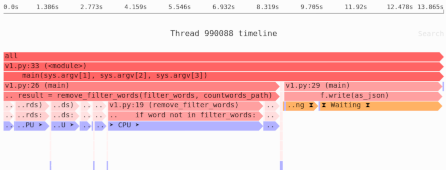
We’ve learned that our naive Pandas implementation used 909MB of memory. That’s a lot! So next we’ll measure the sources of memory usage using the Sciagraph performance and memory profiler.
This report claims 1.2GB of memory allocated; previously we saw that max resident memory was 900MB. The difference is due to measuring different things.
Here’s where most of the memory was allocated:
- 100MB from filtering down to headway rows only.
- 75MB from calculating the median.
- 1000MB in the Arrow library, which is used to load the data. This is not Python code, and Sciagraph doesn’t yet show native callstacks for memory allocations, so it’s not clear exactly which part of loading the data is responsible.
Clearly we want to focus on the last item, but we also have less details there. We could switch to the Memray memory profiler, which does give native (C) callstacks. However, a little thought will suggest at least part of the problem, and the obvious next step. Our current processing involves loading lots of data and then throwing much of it away.
In particular, we:
- Load all the data; this is where Arrow gets involved and allocates a huge amount of memory.
- Drop many rows, specifically those that don’t have headway data.
- Ignore many columns of data which we’re not using in this query.
Chunking or batching is one of the basic techniques for reducing memory usage. If we loaded the data in chunks, rather than all at once, we could filter the data on a chunk by chunk basis. Then we could merge the much-smaller chunks and run our logic on a much smaller amount of data.
Step 4: A more optimized Pandas implementation
Here’s an implementation based on our new insight:
import pandas as pd
import pyarrow.parquet as pq
def find_worst_headways():
# Load the data in chunks:
chunks = []
parquet_file = pq.ParquetFile(
"MBTA-2022-05.parquet"
)
for batch in parquet_file.iter_batches():
chunk = batch.to_pandas()
chunk = chunk[chunk["standard_type"] == "Headway"]
del batch
# Calculate headway ratio:
chunk["headway_ratio"] = (
chunk["headway"] / chunk["scheduled_headway"]
)
# Store the columns we care about for this chunk:
chunks.append(chunk[
["route_id", "direction_id", "headway_ratio"]
])
del parquet_file
# Concatenate into one big DataFrame.
# Not ideal, involves two copies in memory at once...
data = pd.concat(chunks)
del chunks
# Group by route and direction (Inbound/Outbound):
by_route = data.groupby(["route_id", "direction_id"])
# Find median day's headway ratio for each route:
median_headway = by_route[["headway_ratio"]].median()
# Return the worst 5 routes:
return median_headway.nlargest(
5, columns=["headway_ratio"]
)
print(find_worst_headways())
The output is the same, but it uses much less memory:
User time (seconds): 1.10
System time (seconds): 1.26
Percent of CPU this job got: 348%
Elapsed (wall clock) time (h:mm:ss or m:ss): 0:00.68
Maximum resident set size (kbytes): 647316
We’ve gone from 900MB max resident memory to 650MB, a decent improvement.
Step 5: Try swapping out PyArrow for fastparquet
In our original memory profiling we saw that PyArrow was responsible for the bulk of allocated memory, as part of loading the Parquet file.
Pandas can also load Parquet files with a different library called fastparquet, so we can switch both our naive and optimized versions to use that and see how it impacts memory usage.
Here’s what we changed in the naive version:
# ...
data = pd.read_parquet("MBTA-2022-05.parquet",
engine="fastparquet")
# ...
And the optimized version:
import pandas as pd
import fastparquet as pq
def find_worst_headways():
# Load the data in chunks:
chunks = []
parquet_file = pq.ParquetFile("MBTA-2022-05.parquet")
for chunk in parquet_file.iter_row_groups():
# Calculate headway ratio:
chunk["headway_ratio"] = (
chunk["headway"] / chunk["scheduled_headway"]
)
# ...
When measuring memory usage of the resulting code, it turns out that Fastparquet uses far less memory than PyArrow for the naive version. But the optimized version is actually worse! I could spend the time trying to figure out why but that’s probably too much of a digression. At a guess, it’s loading the whole file, and we need to adjust the row groups to be smaller when creating the Parquet file if we want to get any benefit from chunking.
Here’s a summary of our various implementation so far:
| Implementation | Max Resident RAM | Elapsed seconds | CPU seconds |
|---|---|---|---|
| Pandas naive (PyArrow) | 909MB | 0.44 secs | 2.17 secs |
| Pandas optimized (PyArrow) | 650MB | 0.68 secs | 2.36 secs |
| Pandas naive (Fastparquet) | 400MB | 0.71 secs | 2.07 secs |
| Pandas “optimized” (Fastparquet) | 420MB | 0.71 secs | 2.04 secs |
Lazy processing, lazy programmer: using less memory with Polars
So far we’ve learned that a manually-implemented batching implementation can, at least with PyArrow, reduce memory usage in Pandas. Annoyingly, this requires us to manually restructure how the data is represented and loaded. Ideally our library would do that for us, but unfortunately this is not possible when using Pandas.
Pandas is a eager API: you tell it to do something, and it immediately does it. So if you tell it to load a file, it will load all of it into memory; it has no way of knowing you intend to drop half the data on the next line of code.
The alternative is a lazy API that allows you to string together a series of operations—loading, filtering, aggregating, transforming—without actually doing any work. After creating this series of operations, you can then separately tell the library to execute the whole thing.
A smart lazy library can then look at all the operations, and come up with an optimized execution plan that takes into account everything you plan to do—and everything you plan not to do. For example:
- If you’re not touching a column at all, there is no need to load it into memory.
- If batching is possible, the library could do batching for you automatically. Polars lets you explicitly ask for this sort of optimization; see the mention of streaming below.
Polars is an alternative to Pandas with many benefits, like multi-core processing—and it supports both eager and lazy APIs. Using the lazy API can mean lower memory usage without having to do extra work to manually batch data processing.
Our Polars implementation
Polars’ eager loading APIs tend to start with read_*, and the lazy loading APIs start with scan_*.
Here’s what our code looks like when reimplemented with the Polars lazy API, specifically scan_parquet():
import polars as pl
def headways_sorted_worst_first():
# Load the data lazily:
data = pl.scan_parquet("MBTA-2022-05.parquet")
# Filter down to headway points only and then select
# the data we need:
data = data.filter(
pl.col("standard_type") == "Headway"
).select(
[
pl.col("route_id"),
pl.col("direction_id"),
pl.col("headway") / pl.col("scheduled_headway"),
]
)
# Group by route and direction (Inbound/Outbound):
by_route = data.group_by(["route_id", "direction_id"])
# Find median headway ratio for each route:
median_headway = by_route.agg(
pl.col("headway").median()
)
# Get the top 5 worst results:
return median_headway.top_k(
5,
by="headway"
).sort("headway", descending=True)
# Create the query:
query = headways_sorted_worst_first()
# Actually run the query:
result = query.collect()
print(result)
When we were using Pandas, every call did some work.
With Polars’ lazy API, nothing actually happens until we call collect().
At that point, it can use a query planner to come up with an optimized execution strategy.
Only then does Polars do the work of loading, filtering, and aggregating the data.
Future Polars releases might improve the query planner (or, perhaps, make it worse). With Pandas’ eager API, structural optimizations require you to restructure your code; for good or for bad, the execution strategy is up to you.
Memory usage and performance comparison
Let’s see how Polars’ lazy implementation compares to our previous implementations:
| Implementation | Max Resident RAM | Elapsed seconds | CPU seconds |
|---|---|---|---|
| Pandas naive (PyArrow) | 909MB | 0.44 secs | 2.17 secs |
| Pandas optimized (PyArrow) | 650MB | 0.68 secs | 2.36 secs |
| Pandas naive (Fastparquet) | 400MB | 0.71 secs | 2.07 secs |
| Pandas “optimized” (Fastparquet) | 420MB | 0.71 secs | 2.04 secs |
| Polars (lazy) | 250MB | 0.11 secs | 0.44 secs |
Polars uses less memory, it finishes faster, and it uses less CPU. Not bad at all!
Note: This is with Polars 1.33 and Pandas 2.3. When I originally wrote this article Polars used 150MB max resident RAM, so sadly it’s gotten worse. Pandas optimized used to use 360MB, so it has gotten significantly worse too.
Even better, we didn’t have to restructure our code to manually implement batching; either Polars did it for us, or it applied some other memory reduction technique.
And the reason really is the lazy API: merely loading the file with Polars’ eager read_parquet() API results in 310MB max resident RAM.
Note that Polars includes a streaming mode where it specifically tries to use batch APIs to keep memory down.
Just call collect(engine="streaming") instead of collect().
In this case it didn’t meaningfully reduce memory usage, but give it a try if you’re dealing with large files.
Note: Whether or not any particular tool or technique will help depends on where the actual memory bottlenecks are in your software.
Need to identify the memory and performance bottlenecks in your own Python data processing code? Try the Sciagraph profiler, with support for profiling both in development and production macOS and Linux, and with built-in Jupyter support.
Other lazy libraries
There are other libraries that implement lazy APIs:
- Dask’s dataframe support is a wrapper around Pandas; unlike Polars, its API matches Pandas as much as possible while still being lazy. I actually tried Dask with the naive Pandas implementation above and unfortunately it did not save any memory, but I did not spent a lot of time on it so your mileage may vary.
- Vaex is another library implementing a lazy DataFrame, and like Polars it is not compatible with Pandas. I did not try Vaex for this example.
Polars isn’t magic
Just because Polars did better in this case doesn’t mean it always will. No doubt there are also cases where the query planner makes non-optimal decisions.
And it certainly can’t solve all your memory usage problems automatically. Importantly, you still need some manual intervention to make sure you’re starting with efficient data types and an efficient-to-load file format.
When I ran the same code on the original CSVs instead of Parquet files, with strings instead of categoricals or timestamp data types, the optimized Pandas version actually used less memory than Polars. Possibly Python’s string interning compensated for the lack of categoricals. Naively converting the CSV into Parquet wouldn’t have been enough to get memory usage. Replacing strings with categoricals and actual time/date records is important if we want to keep memory usage low.
Keeping that in mind, Polars’ lazy mode is very compelling as a way to process large amounts of data with limited memory—and it’s also both fast and parallelized!